

a.k.a. Bottom-up Mapping
The first step to making this map is to clone and order small pieces of chromosomes which have been cut and the ordered fragments then form contigs (contiguous DNA blocks).
Currently, the resulting library of clones varies in size from 10 000bp to 1Mb.
taken from: http://www.genome.iastate.edy/edu/doe/prim2.html
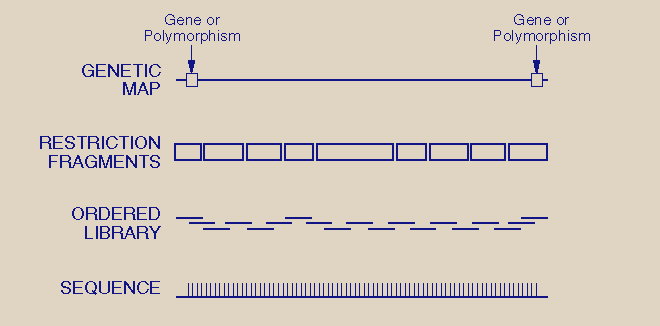 These stable clones that
are produced are made available and accessible to other researchers. Contig
construction can be verified by FISH (see chromosomal map for what FISH is).
These stable clones that
are produced are made available and accessible to other researchers. Contig
construction can be verified by FISH (see chromosomal map for what FISH is).
A complete chromosomal segments is represented by the small library of clones made in the contig maps. This map is useful for finding genes in an area of under 2 Mb (which is a small area), but are difficult to extend over large stretches of a chromosome because not all regions are clonable.
The time consuming process of using DNA probes is a way to fill in the missing gaps, but it not efficient.

Using artificially constructed chromosome vectors that carry human DNA fragments as large as 1Mb, large pieces of DNA can be cloned. The vectors are stored and maintained as artificial chromosomes in yeast cells (YACs). YACs reduce the number of chromosomes that need to be ordered because many YACs are the length of an entire human gene.
The process in which smaller-insert vectors are made from the clone of the fragments of the original insert is called sub cloning. This is a process that will produce a more detailed map of a large YAC insert.
Due to the instability of YAC regions, large-capacity bacterial vectors are also being developed. This will be vectors that can accommodate large inserts.